How Advanced PAT Aids Quality By Digital Design In mRNA Manufacturing
By Adithya Nair, Ph.D., and Zoltán Kis, Ph.D., Department of Biological and Chemical Engineering, University of Sheffield
Quality by digital design (QbDD) is an improvement over the quality by design (QbD) paradigm. Here, computational models are used to characterize, monitor, control, and improve manufacturing processes.
A few ways in which these computational models are implemented include digital twins (DT) and soft sensors. These models can be used for a wide range of activities, including but not limited to measuring system state variables, aiding process development, redundant sensors, advanced process control (predictive control), and real-time process optimization. QbDD implementation, however, is reliant on the quantity and quality of process data. Moreover, the data acquisition needs to be in real time, and provisions for communication between computational models and control components should be provided.
The vital interface between the actual manufacturing process and the computational models is provided by process analytical technology (PAT). Advanced PAT is crucial in implementing real-time monitoring and control of the manufacturing process. QbDD is especially powerful when combined with multi-product manufacturing platform technologies, such as the mRNA platform.
Current Landscape Of The mRNA Manufacturing Platform
The mRNA manufacturing platform is still in the developing phase and lacks the PAT tools available for certain mature biomanufacturing processes. The analysis is mostly offline or at-line and lacks meaningful real-time monitoring or control. However, this is changing as there is a strong demand for establishing a robust platform manufacturing system to meet the growing demand for mRNA-based products. Critical process parameters (CPPs), like pH, ionic strength, and temperature, can be monitored and controlled during mRNA product manufacturing. However, monitoring and controlling underlying reaction kinetics is necessary to improve specific critical quality attributes (CQAs) and key performance indicators (KPIs).
Fluorescent-based and at-line high-performance-liquid-chromatography-based (HPLC) methods have been used to monitor the in vitro transcription reaction and provide some near real-time monitoring and control. Similarly, UV absorbance-based monitoring is an integral part of downstream purification strategies. However, a new set of theorized sensors like NIR and Raman could give further information on substrate utilization (ribonucleotide conversion to mRNA), real-time monitoring of different reaction components (ribonucleotide, mRNA, RNA polymerase, Mg2+), and possibly detection of process and product-related impurities (immunogenic dsRNA).
Challenges: Implementation of advanced PAT is constrained either by a lack of specialized sensors or, in certain cases, appropriate analysis of data generated by available sensors. Unlike conventional chemical synthesis, bioprocesses like mRNA manufacturing generate high-dimensional data that are difficult to acquire and analyze. Moreover, after the analysis is done, there needs to be robust communication between the developed computational models and the manufacturing control components (including actuators) to achieve real-time process control.
Why Does mRNA Manufacturing Matter?
mRNA-based products have seen a stark increase in their potential applications after the success of the two approved mRNA-based vaccines during the SARS-CoV2 pandemic. Apart from their application in prophylactic vaccines, they are also being considered as a potential therapeutic modality. An increase in fiscal investments and research and development as well as clinical trials dedicated to mRNA-based products is evidence of the field’s future growth.1-3
QbD And QbDD Approach In mRNA Manufacturing
Manufacturing mRNA-based products to meet the growing demand requires a well-characterized, robust, and reliable process. Similar to other biopharmaceutical manufacturing processes, the QbD approach could provide the foundation on which a platform mRNA manufacturing system can be built. The QbD approach begins with defining a quality target product profile (QTPP) based on patient requirements. Subsequently, the product’s specific CQAs are identified based on risk assessments yielding severity scores for each quality attribute for both patient safety and product efficacy. Furthermore, the impacts of CPPs within the manufacturing process on the identified CQAs and KPIs are mapped and quantified to reduce the overall risks and improve product quality. A manufacturing design space based on the interactions between the CQAs and CPPs helps establish the optimal ranges for the CPPs that correspond to CQAs within the predetermined quality and regulatory limits. By operating the process with this validated design space, the QbD approach thus ensures that quality is built into the product rather than just tested after the product is manufactured. Moreover, the QbD approach facilitates regulatory approval of future process-related changes as long as the initially approved design space is maintained.4-6
An improvement over QbD is the quality by digital design approach, which utilizes advanced computational models to map and analyze the CQA-CPP interactions. The computational models created in the QbDD paradigm are used for advanced process monitoring and process control. Digital tools such as soft sensors and digital twins are integral to the QbDD approach.7-8
Unlike the conventional QbD approach, however, the QbDD approach is heavily reliant on the quality and quantity of data from the manufacturing process. Extensive data from the manufacturing process is used for the initial development of the computational models and, for their subsequent deployment, a steady data stream from the process is necessary for implementing real-time process monitoring and control. PAT is the interface between the physical manufacturing process and the digital tools employed within the QbDD paradigm.9-10
Current Landscape Of QbDD And Platform mRNA Manufacturing
Currently, mRNA manufacturing processes follow batch operation modes similar to other conventional biopharmaceuticals. The overall manufacturing can be divided into template manufacturing, drug substance manufacturing, drug product manufacturing, and fill/finish operation.10 Figure 1 depicts an mRNA manufacturing process along with its associated unit operations.
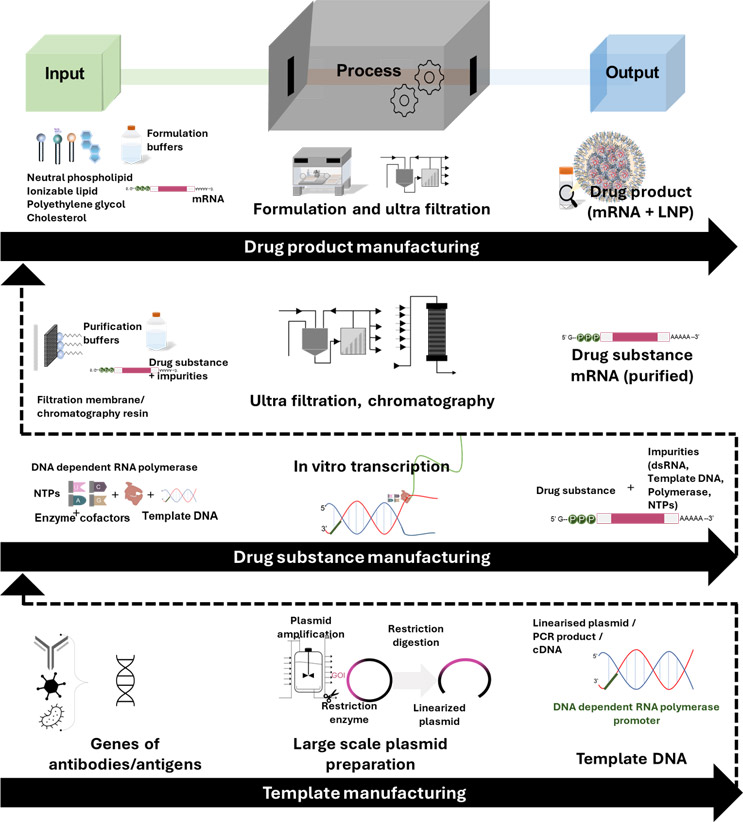
Fig.1. Illustration depicting an mRNA manufacturing system. The overall manufacturing is divided into template manufacturing, drug substance manufacturing (mRNA synthesis and purification), drug product manufacturing (formulated mRNA-based product) and fill-finish operation (mostly outsourced).
Template And Drug Substance Manufacturing
Drug substance manufacturing focuses on mRNA synthesis (although, in some instances, the template DNA synthesis is also included along with this step). mRNA synthesis is done with a cell-free enzymatic process called in vitro transcription (IVT); here, the mRNA is transcribed from a template DNA using an RNA polymerase (enzyme) and ribonucleotides (substrate) along with other reaction components.
The reaction is usually performed at an optimum temperature of 37 degrees C from anywhere between 1 and 4 hours.11, 12 The template DNA from which the mRNA is transcribed can be synthesized in various ways, including PCR amplification, plasmid DNA amplification (combined with a linearization step), or enzymatic synthesis (e.g., doggybone DNA).13, 14
The IVT reaction has been studied in depth, and multiple mechanistic, data-driven, and hybrid models have been proposed to monitor and predict the system state variables to ensure desired quality and process control. The process parameters monitored in the IVT stage normally include the process temperature, pH, and substrate concentration.
Meaningful real-time monitoring and control may be restricted to only process temperature and pH. The PAT involved in this unit operation may be borrowed from other biomanufacturing processes, including but not restricted to microbial fermentation and animal cell culture.15
Substrate monitoring has been reported but remains a near real-time process monitoring and control option at best.16
The purification of synthesized mRNA from the IVT stage is usually achieved with tangential flow filtration and different types of chromatography (ion exchange, size exclusion, multimodal, or affinity chromatography); the sequence of unit operations depends on the manufacturer, but both of these techniques are broadly employed.6, 17
The purification processes transform the synthesized mRNA into the drug substance/active pharmaceutical ingredient. PAT used in the purification stage mainly monitors the concentration of the API before and after the unit operation. For mRNA purification, UV absorbance of the process output is normally used to quantify the mRNA concentration.15, 18
This measurement is usually done with either probe or flow cell-based sensors. A classic limitation of such UV absorbance-based quantification is that it does not differentiate between the mRNA and the unused ribonucleotides from the synthesis stage (process impurities). This limitation may be overcome with advanced analytical techniques such as HPLC, capillary electrophoresis, and mass spectrometry. However, such advanced analytical techniques are generally not integrated with the manufacturing process, and feedback from these techniques cannot be used to control processes.
Drug Product Manufacturing
mRNA-based products are mostly formulated to facilitate their biological uptake (uptake of the large negatively charged mRNA molecules across the lipid bilayer) and improve the overall functionality of the drug product. The mRNA is formulated with either lipids or other transfecting agents, and the most widely studied among these include lipid nanoparticles (LNPs), cationic nanoemulsions (CNEs), cationic peptides, and polymers.19
LNPs, which were used in the formulation of the two approved SARS CoV2 vaccines, can be prepared either with microfluidics or injection methods. These processes are robust and can be scaled successfully to produce consistent products. In either process, an aqueous phase with the mRNA is mixed with an organic phase containing the lipids to produce LNP-encapsulated mRNA.20, 21
CQAs like encapsulation efficiency, LNP size, LNP polydispersity, LNP surface charge, purity, and stability need to be analyzed before the product release. The real-time monitoring of most of these CQAs is difficult, and the technology used for their analysis is too complex to be integrated into the manufacturing system. Newer methods like dynamic light scattering (DLS) have found some applications but are not yet established. CPPs like process temperature and pH have only succeeded with real-time monitoring and control.
Fill/finish And Storage Operations
Fill/finish of the final product is either done in the same manufacturing and formulation facility or outsourced to a dedicated fill/finish facility. Process monitoring and control in these steps are adopted from other biopharmaceutical fill/finish operations and, therefore, remain much more established compared to other unit operations.
The homogeneity of the dispensed product is the most important factor to be considered at this stage. The product CQAs are also affected by exposure to elevated temperatures and the addition of any process-related impurities. The products are also exposed to shear stress that may pose a challenge to product quality. Finally, the filled products need to be stored, and their temperature must be monitored to detect and avoid product degradation from exposure to elevated temperatures.22
Advanced PAT In mRNA Manufacturing
Assessing the status quo, it becomes clear that there is a lag in the development of PAT for the various unit operations within the mRNA manufacturing space and, as mentioned previously, this is detrimental to the implementation of QbDD. Legacy PAT derived from other biopharmaceutical manufacturing is useful to some extent, but it cannot bridge the gap between the present manufacturing processes and those required for bioprocessing 4.0. Even sophisticated analytical technologies (mass spectrometry, RT-PCR, RP-HPLC, etc.) used for various nucleic acid analyses have the disadvantage of being offline or at-line sensors, thus reducing real-time feedback to control and improve the processes. The broader trend in the biopharmaceutical industry is toward more process automation and implementation of QbD rather than conventional manufacturing supported by quality by testing (QbT).9, 23, 24
As with any other biomanufacturing, the magnum opus of mRNA manufacturing will be a well-characterized, well-monitored, automated, continuous process. This goal can be achieved if the CQAs of the API and the formulated product are measured throughout the manufacturing process, including the final storage. Implementing advanced PAT is crucial for developing and deploying soft sensors and digital twins that are instrumental in advanced process monitoring and control. Figure 2 depicts the role of PAT in the wider implementation of QbDD for mRNA manufacturing.
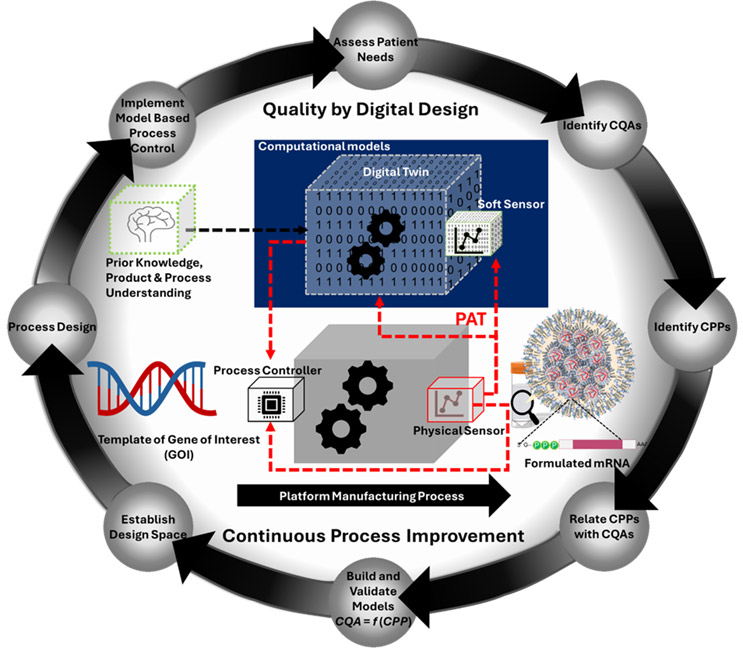
Fig.2. The quality by digital design (QbDD) approach applied to a platform mRNA manufacturing system. It uses computational models to develop advanced monitoring and process control strategies (utilizing soft sensors and digital twins). PAT in this scenario is crucial for process data collection and analysis and communication between controllers and equipment.
Sensor technology advancements and their application in mRNA manufacturing are seeing a definitive change. Sensors that can better monitor CQAs specific to the mRNA field are receiving the necessary attention, and these are being deployed on the drug substance and drug product sides of the manufacturing process. At-line HPLC has been studied to monitor the ribonucleotide concentration during the IVT process and improve the overall yield.
The PAT has helped improve the reaction yield by transitioning to a fed-batch mode of operation. The information was collected at near real-time speeds and used to add the substrate that was being depleted in the reaction. Literature has reported up to 12 g/L yield of the product using this approach, which only requires already established analytical technology.16, 25
Even though the technique uses UV absorbance-based spectra to estimate the product and substrate concentration, the HPLC method helps separate the product from the substrate and quantify the required substrate amount for maintaining the reaction condition toward product formation. The technique has given better insight into substrate utilization and helps with model development, a crucial step in the QbDD approach.18
Proxy soft sensors based on legacy PAT can also help with the development of tools for QbDD; an example of such a scenario is the use of pH probes to monitor reaction kinetics and create feedback for substrate addition. Additionally, there are advancements in legacy PAT that have helped with real-time data collection and communication with controller modules to maintain a robust manufacturing space. UV-Vis spectrum analysis is also a candidate for proxy analysis of the reaction mixture. These can be employed to estimate the system state variables such as substrate concentration and product/process-related impurities. Slight modifications in the reaction mixture by the addition of certain fluorophores are also reported to monitor kinetics.26, 27
Fluorescence resonance energy transfer
Such modifications in the reaction mixture that do not compromise the CQAs of the product and simultaneously facilitate the use of established PAT are highly beneficial and reduce the challenges with novel PAT based on previously unused technology. Fluorescence resonance energy transfer (FRET) was demonstrated to be able to monitor IVT reaction in real time with already available fluorescence detectors. Apart from the established UV-Vis and fluorescent spectral analysis, Raman and IR spectra are also gaining traction as possible PAT for mRNA manufacturing.28
NIR- and Raman-based spectroscopy
NIR and Raman spectra provide molecular fingerprints of the reaction components and could give further information on substrate utilization (ribonucleotide conversion to mRNA), real-time monitoring of different reaction components (ribonucleotide, mRNA, RNA polymerase, Mg2+), and possibly detection of process and product-related impurities (immunogenic dsRNA). The advantages of these spectral analyses include differentiating the product from the substrate without additional sampling or sample preparation. In theory, the Raman and IR spectra are unique for each component of the reaction mixture. They, therefore, should be ideal for monitoring the reaction progress and detecting process/product-related impurities. Moreover, the newer PATs mentioned above are compatible with both the drug substance and drug product stages of mRNA manufacturing based on their wider application in biomanufacturing.28, 29
Challenges In Implementing Advanced PAT
Implementing advanced PAT is constrained either by a lack of specialized sensors or, in certain cases, by the appropriate analysis of data generated by available sensors. Unlike conventional chemical synthesis, bioprocesses like mRNA manufacturing generate high-dimensional data that are difficult to acquire and analyze. Digital tools like soft sensors and DT analyze the data coming from the PAT using different types of computational models. Mechanistic models that are built on mass balance or reaction kinetics are robust and capable of extrapolation, but they are relatively more difficult to develop and computationally complex (leading to lag in real-time feedback). Data-driven models are black boxes that solely rely on process data and are robust within the model input-output variable space (capable of mostly interpolation). These models, however, tend to be faster relative to mechanistic models. A fusion of mechanistic and data-driven models (hybrid models) complements each other’s strengths and mitigates the inherent flaws, making them more suitable for faster analysis of complex data from advanced PAT.30
Moreover, communication between the developed computational models and the manufacturing control components needs to be robust to achieve real-time process control. The data can be obtained manually via the human-machine interface (HMI) or the automated machine-machine interface (MMI) approach. A wider adoption of MMI standards, such as Open Platform Communication (OPC) and OPC Unified Architecture, is crucial for developing and implementing newer monitoring and control strategies.8, 24, 31
Conclusion
mRNA-based products will undoubtedly require a well-characterized and robust manufacturing process as their demand grows in the coming years. Similar to other biopharmaceutical manufacturing, the principles of bioprocess 4.0 and the wider digitization trend will influence the development of this manufacturing process.
Implementing QbDD will be crucial in producing high-quality products, complying with regulatory requirements, and obtaining faster regulatory approval. In the grand scheme of establishing such an automated process, the role of PAT has become more crucial than ever. Advancements in sensor technology, computational models, and standardization of machine-to-machine communication protocols need synergy to achieve digitization in mRNA manufacturing.
References:
- Dutt D, Mazzucato M, Torreele E An mRNA technology transfer programme and economic sustainability in health care. Bulletin of the World Health Organization; Type: Policy & practice 2024.
- Qin S, Tang X, Chen Y, et al (2022) mRNA-based therapeutics: powerful and versatile tools to combat diseases. Signal Transduction and Targeted Therapy 2022 7:1 7:1–35. https://doi.org/10.1038/s41392-022-01007-w
- Damase TR, Sukhovershin R, Boada C, et al (2021) The Limitless Future of RNA Therapeutics. Front Bioeng Biotechnol 9:628137. https://doi.org/10.3389/FBIOE.2021.628137
- U.S. FDA (2009) Guidance for Industry: ICH Q8(R2) Pharmaceutical Development. Workshop: Quality by Design in pharmaceutical
- Rathore AS, Winkle H (2009) Quality by design for biopharmaceuticals. Nature Biotechnology 2009 27:1 27:26–34. https://doi.org/10.1038/nbt0109-26
- Daniel S, Kis Z, Kontoravdi C, Shah N (2022) Quality by Design for enabling RNA platform production processes. Trends Biotechnol 40:1213–1228
- Daniel S, Kis Z, Kontoravdi C, Shah N (2022) A blueprint for quality by digital design to support rapid RNA vaccine process development, manufacturing & supply. Vaccine Insights 01:219–233. https://doi.org/10.18609/VAC.2022.33
- Chen Y, Yang O, Sampat C, et al (2020) Digital Twins in Pharmaceutical and Biopharmaceutical Manufacturing: A Literature Review. Processes 2020, Vol 8, Page 1088 8:1088. https://doi.org/10.3390/PR8091088
- Glassey J, Gernaey K V., Clemens C, et al (2011) Process analytical technology (PAT) for biopharmaceuticals. Biotechnol J 6:369–377. https://doi.org/10.1002/BIOT.201000356
- Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC (2021) mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine 39:2190–2200. https://doi.org/10.1016/J.VACCINE.2021.03.038
- M.J. Tymms, In Vitro Transcription and Translation Protocols, In Vitro Transcription and Translation Protocols (1995). https://doi.org/10.1385/0896032884.
- Beckert B, Masquida B (2011) Synthesis of RNA by in vitro transcription. Methods Mol Biol 703:29–41. https://doi.org/10.1007/978-1-59745-248-9_3
- Karbowniczek K, Rothwell P, Extance J, et al (2017) DoggyboneTM DNA: an advanced platform for AAV production. Cell Gene Ther Insights 3:731–738. https://doi.org/10.18609/cgti.2017.074
- Brunelle JL, Green R (2013) In vitro transcription from plasmid or PCR-amplified DNA. In: Methods in Enzymology. Academic Press Inc., pp 101–114
- Analytical Procedures for mRNA Vaccine Quality (Draft Guidelines)- 2nd Edition | USP-NF. https://www.uspnf.com/notices/analytical-procedures-mrna-vaccines-20230428.
- Skok J, Megušar P, Vodopivec T, et al (2022) Gram-Scale mRNA Production Using a 250-mL Single-Use Bioreactor. Chem Ing Tech 94:1928–1935. https://doi.org/10.1002/cite.202200133
- Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC (2021) mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine 39:2190–2200. https://doi.org/10.1016/J.VACCINE.2021.03.038
- Welbourne EN, Loveday KA, Nair A, et al (2024) Anion exchange HPLC monitoring of mRNA in vitro transcription reactions to support mRNA manufacturing process development. Front Mol Biosci 11: https://doi.org/10.3389/fmolb.2024.1250833
- Ramachandran S, Satapathy SR, Dutta T (2022) Delivery Strategies for mRNA Vaccines. Pharmaceut Med 36:11–20. https://doi.org/10.1007/S40290-021-00417-520.
- Blakney AK, McKay PF, Yus BI, et al (2019) Inside out: optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Therapy 2019 26:9 26:363–372. https://doi.org/10.1038/s41434-019-0095-2
- Streck S, Hong L, Boyd BJ, McDowell A (2019) Microfluidics for the Production of Nanomedicines: Considerations for Polymer and Lipid-based Systems. Pharm Nanotechnol 7:423–443. https://doi.org/10.2174/2211738507666191019154815
- Mirasol F (2021) Meeting Fill/Finish Challenges for COVID-19 Vaccines. Pharmaceutical Technology 2021 Supplement: s22–s23, s28–s22–s23, s28
- Rathore AS, Nikita S, Jesubalan NG (2022) Digitization in bioprocessing: The role of soft sensors in monitoring and control of downstream processing for production of biotherapeutic products. Biosens Bioelectron X 12:100263. https://doi.org/10.1016/J.BIOSX.2022.100263
- Helgers H, Hengelbrock A, Schmidt A, Strube J (2021) Digital Twins for Continuous mRNA Production. Processes 2021, Vol 9, Page 1967 9:1967. https://doi.org/10.3390/PR9111967
- Pregeljc D, Skok J, Vodopivec T, et al (2023) Increasing yield of in vitro transcription reaction with at-line high pressure liquid chromatography monitoring. Biotechnol Bioeng 120:737–747. https://doi.org/10.1002/bit.28299
- Lee KH, Song J, Kim S, et al (2023) Real-time monitoring strategies for optimization of in vitro transcription and quality control of RNA. Front Mol Biosci 10:1229246. https://doi.org/10.3389/FMOLB.2023.1229246
- Li Z, Guo H, Xu F, et al (2019) Sensitive monitoring of RNA transcription by optical amplification of cationic conjugated polymers. Talanta 203:314–321. https://doi.org/10.1016/J.TALANTA.2019.05.052
- Matuszczyk JC, Zijlstra G, Ede D, et al (2023) Raman spectroscopy provides valuable process insights for cell-derived and cellular products. Curr Opin Biotechnol 81:102937. https://doi.org/10.1016/J.COPBIO.2023.102937
- Thakur G, Masampally V, Kulkarni A, Rathore AS (2022) Process Analytical Technology (PAT) Implementation for Membrane Operations in Continuous Manufacturing of mAbs: Model-Based Control of Single-Pass Tangential Flow Ultrafiltration. AAPS Journal 24:1–10. https://doi.org/10.1208/S12248-022-00731-Z
- Gaddem MR, Kim J, Matsunami K, et al (2024) Roles of mechanistic, data-driven, and hybrid modeling approaches for pharmaceutical process design and operation. Curr Opin Chem Eng 44:101019. https://doi.org/10.1016/J.COCHE.2024.101019
- Schmidt A, Helgers H, Lohmann LJ, et al (2022) Process analytical technology as key-enabler for digital twins in continuous biomanufacturing. Journal of Chemical Technology and Biotechnology 97:2336–2346
 About The Authors:
About The Authors:
Adithya Nair is a post-doctoral research associate within the UK-SEA Vax Hub at the University of Sheffield. His work focuses on techno-economic modeling and process development for mRNA-based product manufacturing. He obtained his doctoral degree from Osaka University, Japan, and holds a master’s degree in industrial biotechnology.
 Zoltán Kis is a senior lecturer (associate professor) at the Department of Chemical and Biological Engineering at The University of Sheffield and an honorary lecturer at the Department of Chemical Engineering, Imperial College London. Zoltán is leading a multidisciplinary team that is innovating and digitalizing RNA vaccine and therapeutics production platform technologies. His work is addressing the challenges of producing large volumes of RNA-based vaccines and therapeutics, rapidly, at high quality, and at low cost in a disease-agnostic manner. Zoltán has previously worked as a research associate in the Future Vaccine Manufacturing Hub at Imperial College London. He obtained his Ph.D. in bioengineering from Imperial College London, U.K., and holds an M.Sc. in applied biotechnology and a B.Eng. in chemical with biochemical engineering.
Zoltán Kis is a senior lecturer (associate professor) at the Department of Chemical and Biological Engineering at The University of Sheffield and an honorary lecturer at the Department of Chemical Engineering, Imperial College London. Zoltán is leading a multidisciplinary team that is innovating and digitalizing RNA vaccine and therapeutics production platform technologies. His work is addressing the challenges of producing large volumes of RNA-based vaccines and therapeutics, rapidly, at high quality, and at low cost in a disease-agnostic manner. Zoltán has previously worked as a research associate in the Future Vaccine Manufacturing Hub at Imperial College London. He obtained his Ph.D. in bioengineering from Imperial College London, U.K., and holds an M.Sc. in applied biotechnology and a B.Eng. in chemical with biochemical engineering.
